/*
The MIT License (MIT)
Copyright (c) 2014 Pierre Lindenbaum
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
*/
package com.github.lindenb.jvarkit.tools.biostar;
import java.awt.Insets;
import java.io.BufferedReader;
import java.io.IOException;
import java.io.InputStreamReader;
import java.util.LinkedHashMap;
import java.util.List;
import java.util.Map;
import javax.xml.stream.XMLOutputFactory;
import javax.xml.stream.XMLStreamException;
import javax.xml.stream.XMLStreamWriter;
import com.beust.jcommander.Parameter;
import com.github.lindenb.jvarkit.io.IOUtils;
import com.github.lindenb.jvarkit.util.jcommander.Launcher;
import com.github.lindenb.jvarkit.util.jcommander.Program;
import com.github.lindenb.jvarkit.util.log.Logger;
import com.github.lindenb.jvarkit.util.svg.SVG;
/**
BEGIN_DOC
## Example
```bash
curl -s "http://www.tcoffee.org/Courses/Exercises/saragosa_pb_2010/practicals/practical_2/ex.1.19/file/clustalw.msa" |\
java -jar dist/biostar77288.jar > result.svg
```
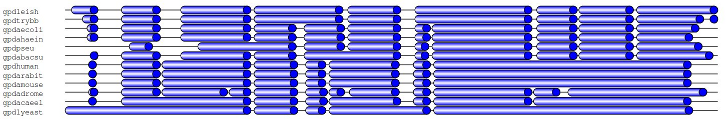
```bash
$ java -jar dist/sam4weblogo.jar IN=in.bam REGION="1:630-719" |\
java -jar dist/biostar77288.jar SEQLOGO=true > result.svg
```
END_DOC
*/
@Program(name=",biostar77288",
description="Low resolution sequence alignment visualization",
biostars=77288,
keywords={"bam","sam","visualization","svg"}
)
public class Biostar77288 extends Launcher
{
private static final Logger LOG= Logger.build(Biostar77288.class).make();
@Parameter(names="-S",description="Input is seqLogo")
private boolean SEQLOGO=false;
@Parameter(names="-W",description=" Alignment width")
private int ALN_WIDTH=1000;
@Parameter(names="-r",description="Use Rect")
private boolean use_rect=false;
private Map<String,Seq> sequences=new LinkedHashMap<String,Seq>();
private int max_length=0;
private int max_name=0;
private int featureHeight=12;
private XMLStreamWriter w=null;
private class Seq
{
String name;
StringBuilder sequence=new StringBuilder();
boolean isGap(int idx)
{
return idx<0 || idx>=sequence.length() || !Character.isLetter(sequence.charAt(idx));
}
}
private void attr(String name,Object o) throws XMLStreamException
{
w.writeAttribute(name, String.valueOf(o));
}
private double base2pix(int n)
{
return (n/(double)this.max_length)*this.ALN_WIDTH + max_name*(double)featureHeight;
}
private void readingClustal(final String IN) throws IOException
{
LOG.info("reading CLUSTALW");
String line;
BufferedReader in=super.openBufferedReader(IN);
while((line=in.readLine())!=null)
{
int ws=0;
if( line.isEmpty() ||
line.startsWith("CLUSTAL W") ||
Character.isWhitespace(line.charAt(0)) ||
(ws=line.indexOf(' '))<1)
{
continue;
}
Seq S=sequences.get(line.substring(0,ws));
if(S==null)
{
S=new Seq();
S.name=line.substring(0,ws);
sequences.put(S.name, S);
}
S.sequence.append(line.substring(ws+1).trim());
max_name=Math.max(S.name.length()+1,max_name);
max_length=Math.max(S.sequence.length(),max_length);
}
in.close();
}
private void readingSeqLogo(String IN) throws IOException
{
LOG.info("reading SeqLogo");
String line;
BufferedReader in=(IN==null?
new BufferedReader(new InputStreamReader(System.in)):
IOUtils.openURIForBufferedReading(IN)
);
while((line=in.readLine())!=null)
{
if( line.isEmpty() ||
!line.startsWith(">"))
{
continue;
}
Seq S=new Seq();
S.name=line.substring(1).trim();
line=in.readLine();
if(line==null) break;
S.sequence.append(line.trim());
sequences.put(S.name, S);
max_name=Math.max(S.name.length()+1,max_name);
max_length=Math.max(S.sequence.length(),max_length);
}
in.close();
}
private int run(String IN) throws IOException
{
try
{
if(this.SEQLOGO)
{
readingSeqLogo(IN);
}
else
{
readingClustal(IN);
}
final Insets svgInset=new Insets(10, 10, 10, 10);
final Insets rowInset=new Insets(7,7,7,7);
XMLOutputFactory xmlfactory= XMLOutputFactory.newInstance();
this.w= xmlfactory.createXMLStreamWriter(stdout(),"UTF-8");
w.writeStartDocument("UTF-8","1.0");
w.writeStartElement("svg");
w.writeDefaultNamespace(SVG.NS);
w.writeAttribute("width", ""+(max_name*featureHeight+1000)+(svgInset.top+svgInset.left));
w.writeAttribute("height", ""+(sequences.size())*(featureHeight+rowInset.top+rowInset.left)+(svgInset.top+svgInset.left));
w.writeAttribute("style", "stroke:none;stroke-width:1px;text-anchor:start;font-size:"+featureHeight+"px;font-family:Courier;");
attr("version", "1.0");
int y=svgInset.top;
w.writeStartElement("defs");
w.writeStartElement("linearGradient");
attr("id","grad");attr("x1","0");attr("x2","0");attr("y1","0");attr("y2","1");
w.writeEmptyElement("stop");attr("offset","0%");attr("stop-color","blue");
w.writeEmptyElement("stop");attr("offset","50%");attr("stop-color","white");
w.writeEmptyElement("stop");attr("offset","100%");attr("stop-color","blue");
w.writeEndElement();
w.writeEndElement();
w.writeCharacters("\n");
for(Seq seq:this.sequences.values())
{
y+=rowInset.top;
w.writeStartElement("g");
w.writeAttribute("transform", "translate("+svgInset.left+","+y+")");
w.writeStartElement("text");
attr("x",0);
attr("y",featureHeight);
w.writeCharacters(seq.name);
w.writeEndElement();
int prev_k=0;
int k1=0;
while(k1< max_length)
{
if(seq.isGap(k1))
{
k1++;
continue;
}
int k2=k1;
while(k2< max_length && !seq.isGap(k2))
{
++k2;
}
w.writeEmptyElement("line");
attr("style","stroke:black;");
attr("x1",base2pix(prev_k));
attr("y1",featureHeight/2);
attr("x2",base2pix(k1));
attr("y2",featureHeight/2);
w.writeEmptyElement("rect");
attr("x",base2pix(k1));
attr("y", "0");
if(!this.use_rect)
{
attr("rx",featureHeight/2.0);
attr("ry",featureHeight/2.0);
}
attr("width",(base2pix(k2)-base2pix(k1)));
attr("height",featureHeight);
attr("style","stroke:black;fill:"+(this.use_rect?"white":"url(#grad)"));
if(!this.use_rect)
{
w.writeEmptyElement("ellipse");
attr("cx",base2pix(k2)-featureHeight/2.0);
attr("cy",featureHeight/2.0);
attr("rx",featureHeight/2.0);
attr("ry",featureHeight/2.0);
attr("style","fill:blue;stroke:black;");
}
k1=k2;
prev_k=k2;
}
w.writeEmptyElement("line");
attr("style","stroke:black;");
attr("x1",base2pix(prev_k));
attr("y1",featureHeight/2);
attr("x2",base2pix(max_length));
attr("y2",featureHeight/2);
w.writeEndElement();
w.writeCharacters("\n");
y+=rowInset.bottom;
}
w.writeEndElement();
w.writeEndDocument();
w.close();
return 0;
}
catch (Exception err)
{
LOG.error(err);
return -1;
}
}
@Override
public int doWork(final List<String> args) {
try {
return run(oneFileOrNull(args));
} catch (IOException e) {
LOG.error(e);
return -1;
}
}
public static void main(String[] args) {
new Biostar77288().instanceMainWithExit(args);
}
}