/*
The MIT License (MIT)
Copyright (c) 2017 Pierre Lindenbaum
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
*/
package com.github.lindenb.jvarkit.tools.bam2graphics;
import java.awt.AlphaComposite;
/**
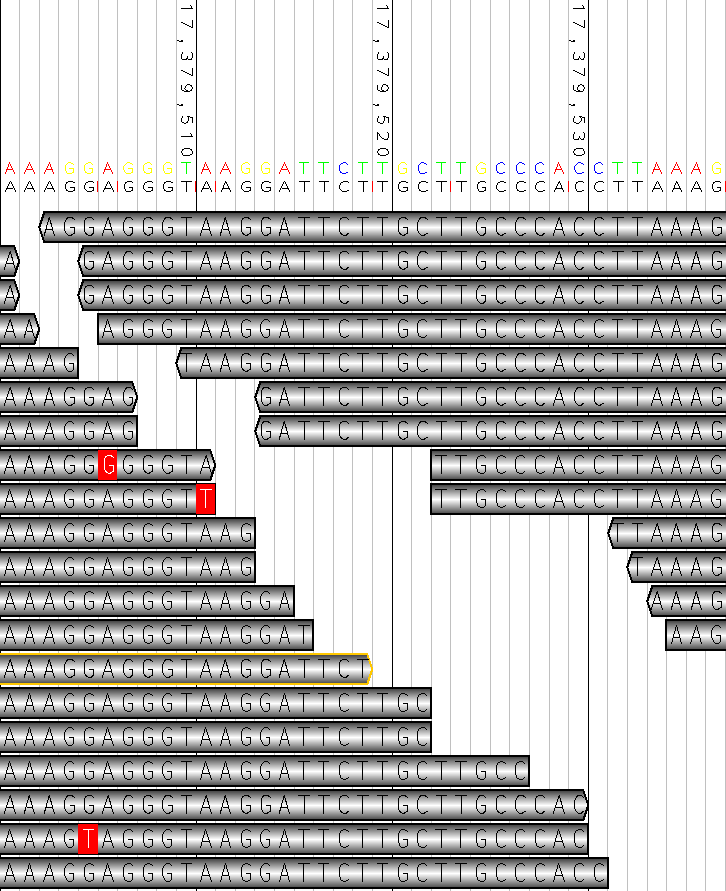
BEGIN_DOC
## Examples
### Example 1
```
java -jar dist/bam2raster.jar \
-o ~/jeter.png \
-r 2:17379500-17379550 \
-R human_g1k_v37.fasta \
sample.bam
```
### Example 2
```
java -jar dist/bam2raster.jar -R ref.fa -r rotavirus:150-200 data/*.bam -o out.png --limit 10 --clip --noReadGradient --highlight 175
```
## Misc
I use the UCSC/IGV color tag 'YC' when available (see also samcolortag)
## Screenshots
<img src="https://raw.github.com/lindenb/jvarkit/master/doc/bam2graphics.png"/>
<img src="https://pbs.twimg.com/media/C_eTeXtW0AAAC-v.jpg"/>
END_DOC
*/
import java.awt.BasicStroke;
import java.awt.Color;
import java.awt.Composite;
import java.awt.Dimension;
import java.awt.Graphics2D;
import java.awt.LinearGradientPaint;
import java.awt.Paint;
import java.awt.RenderingHints;
import java.awt.Shape;
import java.awt.Stroke;
import java.awt.geom.AffineTransform;
import java.awt.geom.GeneralPath;
import java.awt.geom.Line2D;
import java.awt.geom.Rectangle2D;
import java.awt.image.BufferedImage;
import java.io.File;
import java.util.ArrayList;
import java.util.Arrays;
import java.util.HashMap;
import java.util.HashSet;
import java.util.List;
import java.util.Map;
import java.util.Set;
import java.util.TreeMap;
import java.util.function.Function;
import java.util.function.Predicate;
import javax.imageio.ImageIO;
import com.beust.jcommander.Parameter;
import com.github.lindenb.jvarkit.io.IOUtils;
import com.github.lindenb.jvarkit.lang.AbstractCharSequence;
import com.github.lindenb.jvarkit.util.Counter;
import com.github.lindenb.jvarkit.util.Hershey;
import com.github.lindenb.jvarkit.util.bio.IntervalParser;
import com.github.lindenb.jvarkit.util.bio.samfilter.SamFilterParser;
import com.github.lindenb.jvarkit.util.jcommander.Launcher;
import com.github.lindenb.jvarkit.util.jcommander.Program;
import com.github.lindenb.jvarkit.util.log.Logger;
import com.github.lindenb.jvarkit.util.picard.GenomicSequence;
import com.github.lindenb.jvarkit.util.samtools.SAMRecordPartition;
import com.github.lindenb.jvarkit.util.swing.ColorUtils;
import htsjdk.samtools.reference.IndexedFastaSequenceFile;
import htsjdk.samtools.util.Interval;
import htsjdk.variant.variantcontext.VariantContext;
import htsjdk.variant.vcf.VCFFileReader;
import htsjdk.samtools.Cigar;
import htsjdk.samtools.CigarElement;
import htsjdk.samtools.CigarOperator;
import htsjdk.samtools.SAMFileHeader;
import htsjdk.samtools.SamInputResource;
import htsjdk.samtools.SamReader;
import htsjdk.samtools.SamReaderFactory;
import htsjdk.samtools.filter.SamRecordFilter;
import htsjdk.samtools.SAMRecord;
import htsjdk.samtools.SAMRecordIterator;
import htsjdk.samtools.SAMSequenceDictionary;
import htsjdk.samtools.util.CloseableIterator;
import htsjdk.samtools.util.CloserUtil;
/**
BEGIN_DOC
## Example
```
$ java -jar dist/bam2raster.jar -r "scf7180000354095:168-188" \
-o pit.png \
-R scf_7180000354095.fasta scf7180000354095.bam



```
END_DOC
*/
@Program(name="bam2raster",
description="BAM to raster graphics",
keywords={"bam","alignment","graphics","visualization","png"},
biostars=252491
)
public class Bam2Raster extends Launcher
{
private static final Logger LOG = Logger.build(Bam2Raster.class).make();
@Parameter(names={"-o","--output"},description="Output file. Optional . Default: stdout")
private File outputFile = null;
@Parameter(names={"-nobase","--nobase"},description="hide bases")
private boolean hideBases = false;
@Parameter(names={"-r","--region"},description="restrict to that region. REQUIRED",required=true)
private String regionStr = null;
@Parameter(names={"-R","--reference"},description="indexed fasta reference")
private File referenceFile = null;
@Parameter(names={"-w","--width"},description="Image width")
private int WIDTH = 1000 ;
@Parameter(names={"-N","--name"},description="print read name instead of base")
private boolean printName = false;
@Parameter(names={"-srf","--samRecordFilter"},description=SamFilterParser.FILTER_DESCRIPTION,converter=SamFilterParser.StringConverter.class)
private SamRecordFilter samRecordFilter = SamFilterParser.buildDefault();
@Parameter(names={"-minh","--minh"},description="Min. distance between two reads.")
private int minHDistance=2;
@Parameter(names={"-clip","--clip"},description="Show clipping")
private boolean showClip=false;
@Parameter(names={"--limit","--maxrows"},description="Limit number of rows to 'N' lines. negative: no limit.")
private int maxNumberOfRows=-1;
@Parameter(names={"-depth","--depth"},description="Depth size")
private int depthSize=100;
@Parameter(names={"--noReadGradient"},description="Do not use gradient for reads")
private boolean noReadGradient=false;
@Parameter(names={"--highlight"},description="hightligth those positions.",converter=com.beust.jcommander.converters.IntegerConverter.class)
private Set<Integer> highlightPositions = new HashSet<>() ;
@Parameter(names={"--groupby"},description="Group Reads by")
private SAMRecordPartition groupBy=SAMRecordPartition.sample;
@Parameter(names={"--spaceyfeature"},description="number of pixels between features")
private int spaceYbetweenFeatures=4;
@Parameter(names={"-V","--variants","--vcf"},description="VCF files used to fill the position to hightlight with POS")
private List<String> variants=new ArrayList<>();
private enum AlphaHandler {all_opaque,handler1}
@Parameter(names={"--mapqopacity"},description="How to handle the MAPQ/ opacity of the reads.")
private AlphaHandler alpha_handler=AlphaHandler.all_opaque;
public Bam2Raster()
{
}
private static interface Colorizer
{
public Color getColor(SAMRecord rec);
}
/*
private class QualityColorizer implements Colorizer
{
public Color getColor(SAMRecord rec)
{
int f=rec.getMappingQuality();
if(f>255) f=255;
return new Color(f,f,f);
}
}*/
private static class FlagColorizer implements Colorizer
{
public Color getColor(SAMRecord rec)
{
if(!rec.getReadPairedFlag() || rec.getProperPairFlag()) return Color.BLACK;
if(rec.getMateUnmappedFlag()) return Color.BLUE;
if(rec.getDuplicateReadFlag()) return Color.GREEN;
return Color.ORANGE;
}
}
private Interval interval=null;
private IndexedFastaSequenceFile indexedFastaSequenceFile=null;
private int minArrowWidth=2;
private int maxArrowWidth=5;
//private int featureHeight=30;
private final Hershey hersheyFont=new Hershey();
private Colorizer strokeColorizer=new FlagColorizer();
private final Map<String, PartitionImage> key2partition=new TreeMap<>();
private double convertToX(int genomic)
{
return WIDTH*(genomic-interval.getStart())/(double)(interval.getEnd()-interval.getStart()+1);
}
private double left2pixel(final SAMRecord rec)
{
return convertToX(readStart(rec));
}
private double right2pixel(final SAMRecord rec)
{
return convertToX(readEnd(rec));
}
private int readStart(final SAMRecord rec)
{
return showClip?rec.getUnclippedStart():rec.getAlignmentStart();
}
private int readEnd(final SAMRecord rec)
{
return showClip?rec.getUnclippedEnd():rec.getAlignmentEnd();
}
private class PartitionImage
{
final List<List<SAMRecord>> rows = new ArrayList<>();
final String key;
BufferedImage image=null;
PartitionImage(final String key)
{
this.key=key;
}
private void add(final SAMRecord rec)
{
// pileup
for(final List<SAMRecord> row:this.rows)
{
final SAMRecord last=row.get(row.size()-1);
if(right2pixel(last)+ Bam2Raster.this.minHDistance > left2pixel(rec)) continue;
row.add(rec);
return;
}
if(rec!=null)
{
final List<SAMRecord> row=new ArrayList<SAMRecord>();
row.add(rec);
this.rows.add(row);
}
}
private void build()
{
final Function<SAMRecord,Color> samRecord2color = new ColorUtils.SAMRecordColorExtractor();
final Function<SAMRecord,Float> samRecord2alpha = R->{
final int mapq = R.getMappingQuality();
switch(alpha_handler)
{
case handler1:
if( mapq == SAMRecord.UNKNOWN_MAPPING_QUALITY) return 0.5f;
if( mapq >= 60 ) return 1f;
if(mapq >50 && mapq <= 60) return 0.9f;
if(mapq >40 && mapq <= 50) return 0.8f;
if(mapq >30 && mapq <= 40) return 0.7f;
if(mapq >20 && mapq <= 30) return 0.6f;
if(mapq >10 && mapq <= 20) return 0.5f;
return 0.4f;
case all_opaque: return 1f;
default: return 1f;
}
};
final Function<Character, Color> base2color = C ->
{
switch(Character.toUpperCase(C))
{
case 'N': return Color.BLACK;
case 'A': return Color.RED;
case 'T': return Color.GREEN;
case 'G': return Color.YELLOW;
case 'C': return Color.BLUE;
default: return Color.ORANGE;
}
};
final Predicate<Integer> inInterval = refPos-> !(refPos< Bam2Raster.this.interval.getStart() || refPos> Bam2Raster.this.interval.getEnd());
String positionFormat="%,d";
final int refw=(int)Math.max(1.0, WIDTH/(double)(1+interval.getEnd()-interval.getStart()));
final int ruler_height=String.format(positionFormat,Bam2Raster.this.interval.getEnd()).length()*refw;
//final int margin_top=10+(refw*3)+ruler_height;
final Dimension imageSize=new Dimension(WIDTH,
refw+ Bam2Raster.this.spaceYbetweenFeatures + //contig name
ruler_height + Bam2Raster.this.spaceYbetweenFeatures + //position
refw + Bam2Raster.this.spaceYbetweenFeatures + //ref seq
refw + Bam2Raster.this.spaceYbetweenFeatures + //consensus
(Math.max(0, Bam2Raster.this.depthSize))+(Bam2Raster.this.depthSize>0?Bam2Raster.this.spaceYbetweenFeatures:0)+//depth
(Bam2Raster.this.maxNumberOfRows<0?this.rows.size():Math.min(this.rows.size(), Bam2Raster.this.maxNumberOfRows))*(Bam2Raster.this.spaceYbetweenFeatures+refw)+Bam2Raster.this.spaceYbetweenFeatures
);
this.image = new BufferedImage(
imageSize.width,
imageSize.height,
BufferedImage.TYPE_INT_RGB
);
final CharSequence genomicSequence;
if(Bam2Raster.this.indexedFastaSequenceFile !=null)
{
genomicSequence=new GenomicSequence(
Bam2Raster.this.indexedFastaSequenceFile,
Bam2Raster.this.interval.getContig());
}
else
{
genomicSequence=new AbstractCharSequence()
{
@Override
public int length()
{
return interval.getEnd()+10;
}
@Override
public char charAt(int index)
{
return 'N';
}
};
}
final Graphics2D g= this.image.createGraphics();
g.setRenderingHint(
RenderingHints.KEY_RENDERING,
RenderingHints.VALUE_RENDER_QUALITY
);
g.setColor(Color.WHITE);
g.fillRect(0, 0, imageSize.width, imageSize.height);
LOG.info("image : "+imageSize.width+"x"+imageSize.height);
Map<Integer, Counter<Character>> ref2consensus=new HashMap<Integer, Counter<Character>>();
//draw bases positions
// paint hightlight bckg
for(final Integer refpos: Bam2Raster.this.highlightPositions) {
g.setColor(new Color(255,235,246));
g.fill(new Rectangle2D.Double(
convertToX(refpos),
0,
refw,
this.image.getHeight()
));
}
int y=0;
//print name
{
String title= interval.getContig()+" "+this.key;
g.setColor(Color.BLACK);
hersheyFont.paint(g,
title,
new Rectangle2D.Double(
1,1,
title.length()*refw,
refw
)
);
y+= refw + Bam2Raster.this.spaceYbetweenFeatures;
}
for(int x=Bam2Raster.this.interval.getStart();x<=Bam2Raster.this.interval.getEnd();++x)
{
final double oneBaseWidth=convertToX(x+1)-convertToX(x);
//draw vertical line
g.setColor(x%10==0?Color.BLACK:Color.LIGHT_GRAY);
g.draw(new Line2D.Double(convertToX(x), 0, convertToX(x), imageSize.height));
if((x-Bam2Raster.this.interval.getStart())%10==0)
{
g.setColor(Color.BLACK);
final String xStr=String.format(positionFormat,x);
final AffineTransform tr=g.getTransform();
final AffineTransform tr2=new AffineTransform(tr);
tr2.translate(convertToX( x + 1 ), y);
tr2.rotate(Math.PI/2.0);
g.setTransform(tr2);
hersheyFont.paint(g,
xStr,
0,
0,
ruler_height,
oneBaseWidth
);
g.setTransform(tr);
}
}
y+= ruler_height + Bam2Raster.this.spaceYbetweenFeatures;
// draw ref bases
for(int x=Bam2Raster.this.interval.getStart();x<=Bam2Raster.this.interval.getEnd();++x)
{
final double oneBaseWidth=convertToX(x+1)-convertToX(x);
//paint genomic sequence
final char c=x<1 || x > genomicSequence.length() ? 'N':genomicSequence.charAt(x-1);
g.setColor(base2color.apply(c));
Bam2Raster.this.hersheyFont.paint(g,
String.valueOf(c),
convertToX(x)+1,
y,
oneBaseWidth-2,
oneBaseWidth-2
);
}
y+= refw + Bam2Raster.this.spaceYbetweenFeatures;
// draw consensus here
final int consensus_y = y;
y+= refw + Bam2Raster.this.spaceYbetweenFeatures;
final int depth_y = y;
final int depth_array[]=new int[1+(interval.getEnd()-interval.getStart())];
if(depthSize>0)
{
Arrays.fill(depth_array, 0);
y+= depthSize+Bam2Raster.this.spaceYbetweenFeatures;
}
// draw reads
for(int rowIndex=0;rowIndex < rows.size();++rowIndex)
{
final List<SAMRecord> row = rows.get(rowIndex);
boolean printThisRow= (Bam2Raster.this.maxNumberOfRows <0 || rowIndex < Bam2Raster.this.maxNumberOfRows );
for(final SAMRecord rec:row)
{
final Set<Integer> refposOfInsertions = new HashSet<>();
double x0 = left2pixel(rec);
double x1 = right2pixel(rec);
double y0 = y;
double y1 = y0 + refw;
Shape shapeRec=null;
if(x1-x0 < Bam2Raster.this.minArrowWidth)
{
shapeRec=new Rectangle2D.Double(x0, y0, x1-x0, y1-y0);
}
else
{
final GeneralPath path=new GeneralPath();
double arrow=Math.max(Bam2Raster.this.minArrowWidth,Math.min(Bam2Raster.this.maxArrowWidth, x1-x0));
if(!rec.getReadNegativeStrandFlag())
{
path.moveTo(x0, y0);
path.lineTo(x1-arrow,y0);
path.lineTo(x1,(y0+y1)/2);
path.lineTo(x1-arrow,y1);
path.lineTo(x0,y1);
}
else
{
path.moveTo(x0+arrow, y0);
path.lineTo(x0,(y0+y1)/2);
path.lineTo(x0+arrow,y1);
path.lineTo(x1,y1);
path.lineTo(x1,y0);
}
path.closePath();
shapeRec=path;
}
final Composite oldComposite = g.getComposite();
if(printThisRow) {
g.setComposite(AlphaComposite.getInstance(AlphaComposite.SRC_OVER,samRecord2alpha.apply(rec)));
Color ycColor = samRecord2color.apply(rec);
final Stroke oldStroke = g.getStroke();
g.setStroke(new BasicStroke(2f));
if(noReadGradient) {
if(ycColor==null) ycColor=new Color(255,222,173);
g.setColor(ycColor);
g.fill(shapeRec);
}
else
{
if(ycColor==null) ycColor=Color.DARK_GRAY;
final Paint oldpaint=g.getPaint();
final LinearGradientPaint gradient=new LinearGradientPaint(
0f, (float)shapeRec.getBounds2D().getY(),
0f, (float)shapeRec.getBounds2D().getMaxY(),
new float[]{0f,0.5f,1f},
new Color[]{ycColor,Color.WHITE,ycColor}
);
g.setPaint(gradient);
g.fill(shapeRec);
g.setPaint(oldpaint);
}
g.setColor(Bam2Raster.this.strokeColorizer.getColor(rec));
g.draw(shapeRec);
g.setStroke(oldStroke);
}
final Shape oldClip=g.getClip();
g.setClip(shapeRec);
final Cigar cigar=rec.getCigar();
if(cigar!=null)
{
final Function<Integer,Character> readBaseAt= IDX -> {
final byte bases[]=rec.getReadBases();
if(SAMRecord.NULL_SEQUENCE.equals(bases)) return 'N';
if(IDX<0 || IDX>=bases.length) return 'N';
return (char)bases[IDX];
};
final Function<Integer,Character> readNameAt= readpos -> {
char c1;
if(readpos<rec.getReadName().length())
{
c1=rec.getReadName().charAt(readpos);
c1=rec.getReadNegativeStrandFlag()?
Character.toLowerCase(c1):Character.toUpperCase(c1);
}
else
{
c1=' ';
}
return c1;
};
int refpos= rec.getUnclippedStart();
int readpos=0;
for(final CigarElement ce:cigar.getCigarElements())
{
switch(ce.getOperator())
{
case S:
case H:
{
if(Bam2Raster.this.showClip)
{
g.setColor(Color.PINK);
if(printThisRow) g.fill(new Rectangle2D.Double(
convertToX(refpos),
y0,
convertToX(refpos+ce.getLength())-convertToX(refpos),
y1-y0
));
if(ce.getOperator().equals(CigarOperator.S))
{
final double mutW=convertToX(refpos+1)-convertToX(refpos);
for(int i=0;i< ce.getLength();++i)
{
if(!inInterval.test(refpos+i)) continue;
char c1=readBaseAt.apply(readpos+i);
g.setColor(base2color.apply(c1));
final Shape mut= new Rectangle2D.Double(
convertToX(refpos+i),
y0,
mutW,
y1-y0
);
if( Bam2Raster.this.printName) c1=readNameAt.apply(readpos+i);
if(printThisRow) Bam2Raster.this.hersheyFont.paint(g,String.valueOf(c1),mut);
}
}
}
refpos+=ce.getLength();
if(ce.getOperator().equals(CigarOperator.S)) readpos+=ce.getLength();
break;
}
case I:
{
refposOfInsertions.add(refpos);
readpos+=ce.getLength();
break;
}
case P: break;
case D:
case N:
{
g.setColor(Color.ORANGE);
if(printThisRow) g.fill(new Rectangle2D.Double(
convertToX(refpos),
y0,
convertToX(refpos+ce.getLength())-convertToX(refpos),
y1-y0
));
refpos+=ce.getLength();
break;
}
case EQ:
case X:
case M:
{
for(int i=0;i< ce.getLength();++i)
{
boolean drawbase=!Bam2Raster.this.hideBases;
char c1=readBaseAt.apply(readpos);
/* handle consensus */
Counter<Character> consensus=ref2consensus.get(refpos);
if(consensus==null)
{
consensus=new Counter<Character>();
ref2consensus.put(refpos,consensus);
}
consensus.incr(Character.toUpperCase(c1));
char c2=genomicSequence.charAt(refpos-1);
double mutW=convertToX(refpos+1)-convertToX(refpos);
g.setColor(Color.BLACK);
final Shape mut= new Rectangle2D.Double(
convertToX(refpos),
y0,
mutW,
y1-y0
);
if(ce.getOperator()==CigarOperator.X ||
(c2!='N' && c2!='n' &&
Character.toUpperCase(c1)!=Character.toUpperCase(c2)))
{
drawbase=true;
g.setColor(Color.RED);
if(printThisRow) g.fill(mut);
g.setColor(Color.WHITE);
}
else
{
g.setColor(base2color.apply(c1));
}
//print read name instead of base
if(Bam2Raster.this.printName)
{
drawbase=true;
c1= readNameAt.apply(readpos);
}
if(!inInterval.test(refpos))
{
drawbase=false;
}
if(!printThisRow)
{
drawbase=false;
}
if(drawbase)
{
Bam2Raster.this.hersheyFont.paint(g,String.valueOf(c1),mut);
}
if(inInterval.test(refpos)) {
depth_array[refpos-Bam2Raster.this.interval.getStart()]++;
}
readpos++;
refpos++;
}
break;
}
default: LOG.error("cigar element not handled:"+ce.getOperator());break;
}
}
}
// paint insertions
for(final Integer refpos: refposOfInsertions) {
g.setColor(Color.GREEN);
if(printThisRow) g.fill(new Rectangle2D.Double(
convertToX(refpos),
y0,
2,
y1-y0
));
}
g.setClip(oldClip);
g.setComposite(oldComposite);
}
if(printThisRow)
{
y+=refw+Bam2Raster.this.spaceYbetweenFeatures;
}
}
//print consensus
for(int x=Bam2Raster.this.interval.getStart();x<=Bam2Raster.this.interval.getEnd() ;++x)
{
Counter<Character> cons=ref2consensus.get(x);
if(cons==null || cons.getCountCategories()==0)
{
continue;
}
final double oneBaseWidth=(convertToX(x+1)-convertToX(x))-1;
double x0=convertToX(x)+1;
for(final Character c:cons.keySetDecreasing())
{
double weight=oneBaseWidth*(cons.count(c)/(double)cons.getTotal());
g.setColor(Color.BLACK);
if(genomicSequence!=null &&
Character.toUpperCase(genomicSequence.charAt(x-1))!=Character.toUpperCase(c))
{
g.setColor(Color.RED);
}
hersheyFont.paint(g,
String.valueOf(c),
x0,
consensus_y,
weight,
oneBaseWidth-2
);
x0+=weight;
}
}
// print depth
if(Bam2Raster.this.depthSize>0)
{
double minDepth = Arrays.stream(depth_array).min().orElse(0);
double maxDepth = Arrays.stream(depth_array).max().orElse(1);
if(minDepth==maxDepth) minDepth--;
for(int i=0;i< depth_array.length;++i)
{
final double d= depth_array[i];
final double h= ((d-minDepth)/(maxDepth-minDepth))*Bam2Raster.this.depthSize;
final Rectangle2D.Double rd= new Rectangle2D.Double();
rd.x= convertToX(interval.getStart()+i);
rd.y= depth_y + Bam2Raster.this.depthSize - h;
rd.width = refw;
rd.height = h;
g.setColor(d < 10?Color.RED:(d<50?Color.BLUE:Color.GREEN));
g.fill(rd);
g.setColor(Color.BLACK);
g.draw(rd);
}
final String label="Depth ["+(int)minDepth+" - "+(int)maxDepth+"]";
for(int x=0;x<2;++x) {
g.setColor(x==0?Color.WHITE:Color.BLACK);
hersheyFont.paint(g,
label,
new Rectangle2D.Double(
1+x,
depth_y +x + Bam2Raster.this.depthSize-refw,
label.length()*refw,
refw
)
);
}
}
// paint hightlight
for(final Integer refpos: Bam2Raster.this.highlightPositions) {
g.setColor(Color.RED);
g.draw(new Rectangle2D.Double(
convertToX(refpos),
0,
refw,
this.image.getHeight()
));
}
g.dispose();
}
}
private void scan(final SamReader r) {
final SAMRecordIterator iter=r.query(
interval.getContig(),
interval.getStart(),
interval.getEnd(),
false
);
while(iter.hasNext())
{
final SAMRecord rec=iter.next();
if(rec.getReadUnmappedFlag()) continue;
if(this.samRecordFilter.filterOut(rec)) continue;
if(!this.interval.getContig().equals(rec.getReferenceName())) continue;
if(readEnd(rec) < this.interval.getStart())
{
continue;
}
if(readStart(rec) > this.interval.getEnd()) {
break;
}
final String group=this.groupBy.apply(rec.getReadGroup());
if(group==null) continue;
PartitionImage partition = this.key2partition.get(group);
if( partition == null)
{
partition=new PartitionImage(group);
this.key2partition.put(group,partition);
}
partition.add(rec);
}
iter.close();
}
@Override
public int doWork(final List<String> args) {
if(this.regionStr==null)
{
LOG.error("Region was not defined.");
return -1;
}
if(this.WIDTH<100)
{
LOG.info("adjusting WIDTH to 100");
this.WIDTH=100;
}
SamReader samFileReader=null;
try
{
final SamReaderFactory srf = super.createSamReaderFactory();
if(this.referenceFile!=null)
{
LOG.info("loading reference");
this.indexedFastaSequenceFile=new IndexedFastaSequenceFile(this.referenceFile);
srf.referenceSequence(this.referenceFile);
}
final IntervalParser intervalParser = new IntervalParser(this.indexedFastaSequenceFile==null?null:this.indexedFastaSequenceFile.getSequenceDictionary()).
setFixContigName(true);
this.interval = intervalParser.parse(this.regionStr);
if(this.interval==null)
{
LOG.error("Cannot parse interval "+regionStr+" or chrom doesn't exists in sam dictionary.");
return -1;
}
LOG.info("Interval is "+this.interval );
for(final String vcfFile: IOUtils.unrollFiles(variants)) {
final VCFFileReader vcfFileReader = new VCFFileReader(new File(vcfFile),true);
final CloseableIterator<VariantContext> r=vcfFileReader.query(this.interval.getContig(), this.interval.getStart(), this.interval.getEnd());
while(r.hasNext())
{
this.highlightPositions.add(r.next().getStart());
}
r.close();
vcfFileReader.close();
}
for(final String bamFile: IOUtils.unrollFiles(args))
{
samFileReader = srf.open(SamInputResource.of(bamFile));
final SAMFileHeader header=samFileReader.getFileHeader();
final SAMSequenceDictionary dict=header.getSequenceDictionary();
if(dict==null) {
LOG.error("no dict in "+bamFile);
return -1;
}
if(dict.getSequence(this.interval.getContig())==null){
LOG.error("no such chromosome in "+bamFile+" "+this.interval);
return -1;
}
scan(samFileReader);
samFileReader.close();
samFileReader=null;
}
if(this.key2partition.isEmpty())
{
LOG.error("No data was found.(not Read-Group specified ?");
return -1;
}
this.key2partition.values().stream().forEach(P->P.build());
//assemble everything
int image_width= this.key2partition.values().stream().mapToInt(P->P.image.getWidth()).max().getAsInt();
int image_height= this.key2partition.values().stream().mapToInt(P->P.image.getHeight()).sum();
final BufferedImage img= new BufferedImage(image_width, image_height, BufferedImage.TYPE_INT_RGB);
final Graphics2D g=img.createGraphics();
g.setRenderingHint(
RenderingHints.KEY_RENDERING,
RenderingHints.VALUE_RENDER_QUALITY
);
int y=0;
for(final String key:this.key2partition.keySet()) {
BufferedImage subImg = this.key2partition.get(key).image;
g.drawImage(subImg,0,y,null);
y+=subImg.getHeight();
}
g.dispose();
if(this.outputFile==null)
{
ImageIO.write(img, "PNG", stdout());
}
else
{
LOG.info("saving to "+this.outputFile);
final String format=(this.outputFile.getName().toLowerCase().endsWith(".png")?"PNG":"JPG");
ImageIO.write(img,format, this.outputFile);
}
return RETURN_OK;
}
catch(Exception err)
{
LOG.error(err);
return -1;
}
finally
{
CloserUtil.close(indexedFastaSequenceFile);
CloserUtil.close(samFileReader);
indexedFastaSequenceFile=null;
}
}
public static void main(String[] args)
{
new Bam2Raster().instanceMainWithExit(args);
}
}